VCF Viewer is an extremely small utility which currently only has one purpose: to view the contents of VCF files. A typical vCard VCF file is generally used by popular email clients and for mobile phones.
- Vcard file viewer supports all the editions of vCard such as 4.1, 2.1, iCard, hCard etc. With the help of free viewer, you can easily multiple files at a time. Now, VCF files are available in bulk.
- This viewer allows you to view and print VCF (vCard) files, as well as convert a VCF file to CSV spreadsheet formats. It is easy to use, With just a few clicks, you can get the converted file, and View / Print a VCF file from your browser directly. You can open a VCF file from local computer, Google Drive, Dropbox, and Box.
- 4 View VCF file using vCard Editor A dedicated free VCF file viewer software is another way to open and view contacts from VCF files. There are some options available on the internet.
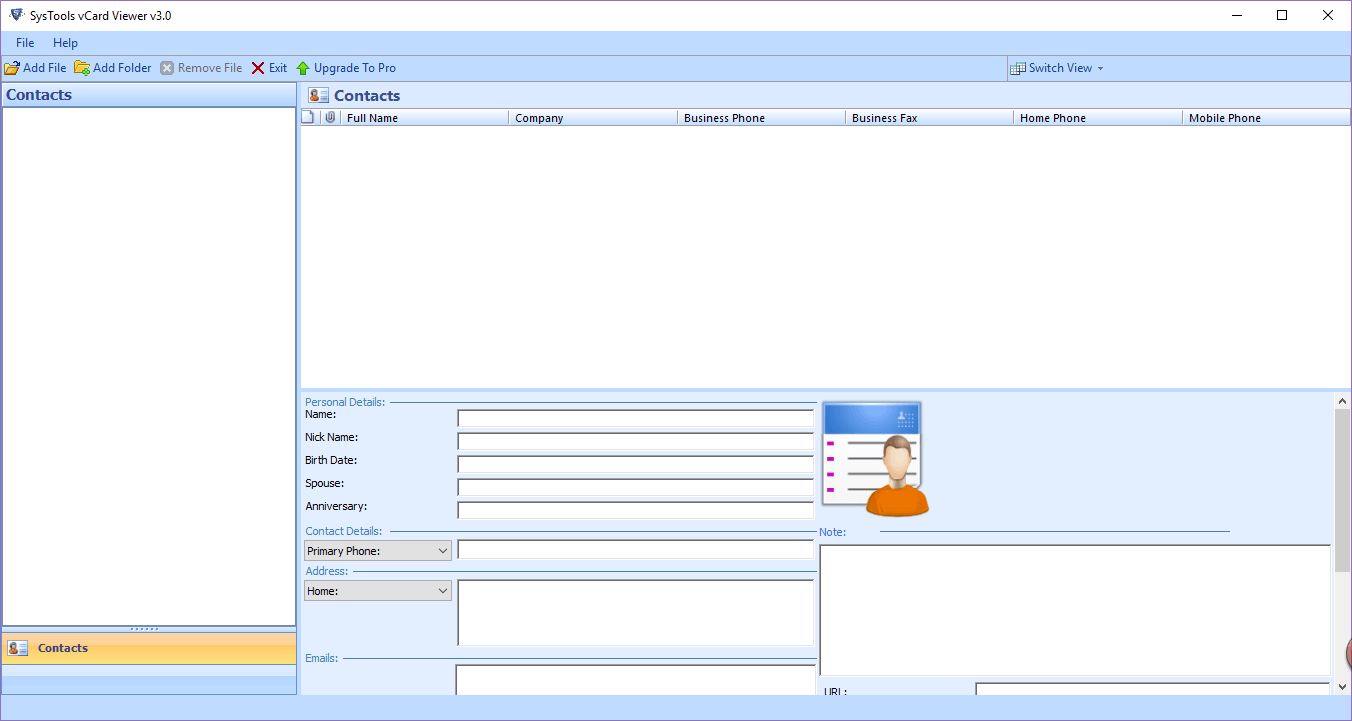
The program itself views the contents of VCF files which have been placed in a directory and finding the information only involves entering the source folder and then scanning. The interface of VCF Viewer will display a list of VCF files in a variety of columns.
Apart from that, VCF Viewer is only a view and is not a vCard editor.
VCF Viewer 1.1 on 32-bit and 64-bit PCs
This download is licensed as freeware for the Windows (32-bit and 64-bit) operating system on a laptop or desktop PC from organizer and pim software without restrictions. VCF Viewer 1.1 is available to all software users as a free download for Windows. As an open source project, you are free to view the source code and distribute this software application freely.
Subsequent downloads and use in other programs and online resources. Select Import VCARD File (.vcf) and click Next. You can open.VCF files as special programs from our list, and the standard 'Address Book' Windows. There is a problem when importing Google Android phonebook backup files to Outlook.The built-in vcf viewer in Windows7.
Filed under:- VCF Viewer Download
- Freeware Organizer and PIM Software
- Open source and GPL software
- Portable Software
- VCF File Viewing Software
Please note that this page describes version 4.0 of the VCF specification. The VCF specification is now maintained by GA4GH. Details of the current version can be found with the HTS specifications.
Please see VCF_4.0_sv for the conventions and extensions adopted by the 1000 Genomes Project for encoding structural variations in VCF 4.0 format.
0. Example
VCF is a text file format (most likely stored in a compressed manner). It contains meta-information lines, a header line, and then data lines each containing information about a position in the genome.
There is an option whether to contain genotype information on samples for each position or not.
Example:
This example shows in order a good simple SNP, a possible SNP that has been filtered out because its quality is below 10, a site at which two alternate alleles are called, with one of them (T) being ancestral (possibly a reference sequencing error), a site that is called monomorphic reference (i.e. with no alternate alleles), and a microsatellite with two alternative alleles, one a deletion of 3 bases (TCT), and the other an insertion of one base (A). Genotype data are given for three samples, two of which are phased and the third unphased, with per sample genotype quality, depth and haplotype qualities (the latter only for the phased samples) given as well as the genotypes. The microsatellite calls are unphased.
1. Meta-information lines
Vcf File Viewer Online
File meta-information is included after the ## string, often as key=value pairs.
The ‘fileformat’ field is always required and should detail the VCF format version number. For example, for VCF version 4.0, this line should read:
##fileformat=VCFv4.0
It is strongly encouraged that information lines describing the INFO, FILTER and FORMAT entries used in the body of the VCF file be included in the meta-information section. Although they are optional, if these lines are present then they must be completely well-formed.
INFO fields should be described as follows (all keys are required):
##INFO=<ID=ID,Number=number,Type=type,Description=”description”>
Possible Types for INFO fields are: Integer, Float, Flag, Character, and String.
The Number entry is an Integer that describes the number of values that can be included with the INFO field. For example, if the INFO field contains a single number, then this value should be 1. However, if the INFO field describes a pair of numbers, then this value should be 2 and so on. If the number of possible values varies, is unknown, or is unbounded, then this value should be ‘.’. Possible Types are: Integer, Float, Character, String and Flag. The ‘Flag’ type indicates that the INFO field does not contain a Value entry, and hence the Number should be 0 in this case. The Description value must be surrounded by double-quotes.
FILTERs that have been applied to the data should be described as follows:
##FILTER=<ID=ID,Description=”description”>
Likewise, Genotype fields specified in the FORMAT field should be described as follows:
##FORMAT=<ID=ID,Number=number,Type=type,Description=”description”>
Possible Types for FORMAT fields are: Integer, Float, Character, and String.
2. The header line syntax
The header line names the 8 fixed, mandatory columns. These columns are as follows:
- #CHROM
- POS
- ID
- REF
- ALT
- QUAL
- FILTER
- INFO
If genotype data is present in the file, these are followed by a FORMAT column header, then an arbitrary number of sample IDs. The header line is tab-delimited.
3. Data lines
Fixed fields
There are 8 fixed fields per record. All data lines are tab-delimited. In all cases, missing values are specified with a dot (“.”). Fixed fields are:
- CHROM chromosome: an identifier from the reference genome. All entries for a specific CHROM should form a contiguous block within the VCF file.(Alphanumeric String, Required)
- POS position: The reference position, with the 1st base having position 1. Positions are sorted numerically, in increasing order, within each reference sequence CHROM. (Integer, Required)
- ID semi-colon separated list of unique identifiers where available. If this is a dbSNP variant it is encouraged to use the rs number(s). No identifier should be present in more than one data record. If there is no identifier available, then the missing value should be used. (Alphanumeric String)
- REF reference base(s): Each base must be one of A,C,G,T,N. Bases should be in uppercase. Multiple bases are permitted. The value in the POS field refers to the position of the first base in the String. For InDels, the reference String must include the base before the event (which must be reflected in the POS field). (String, Required).
- ALT comma separated list of alternate non-reference alleles called on at least one of the samples. Options are base Strings made up of the bases A,C,G,T,N, or an angle-bracketed ID String (”<ID>“). If there are no alternative alleles, then the missing value should be used. Bases should be in uppercase. (Alphanumeric String; no whitespace, commas, or angle-brackets are permitted in the ID String itself)
- QUAL phred-scaled quality score for the assertion made in ALT. i.e. give -10log_10 prob(call in ALT is wrong). If ALT is ”.” (no variant) then this is -10log_10 p(variant), and if ALT is not ”.” this is -10log_10 p(no variant). High QUAL scores indicate high confidence calls. Although traditionally people use integer phred scores, this field is permitted to be a floating point to enable higher resolution for low confidence calls if desired. (Numeric)
- FILTER filter: PASS if this position has passed all filters, i.e. a call is made at this position. Otherwise, if the site has not passed all filters, a semicolon-separated list of codes for filters that fail. e.g. “q10;s50” might indicate that at this site the quality is below 10 and the number of samples with data is below 50% of the total number of samples. “0” is reserved and should not be used as a filter String. If filters have not been applied, then this field should be set to the missing value. (Alphanumeric String)
- INFO additional information: (Alphanumeric String) INFO fields are encoded as a semicolon-separated series of short keys with optional values in the format: <key>=<data>[,data]. Arbitrary keys are permitted, although the following sub-fields are reserved (albeit optional):
- AA ancestral allele
- AC allele count in genotypes, for each ALT allele, in the same order as listed
- AF allele frequency for each ALT allele in the same order as listed: use this when estimated from primary data, not called genotypes
- AN total number of alleles in called genotypes
- BQ RMS base quality at this position
- CIGAR cigar string describing how to align an alternate allele to the reference allele
- DB dbSNP membership
- DP combined depth across samples, e.g. DP=154
- END end position of the variant described in this record (esp. for CNVs)
- H2 membership in hapmap2
- MQ RMS mapping quality, e.g. MQ=52
- MQ0 Number of MAPQ 0 reads covering this record
- NS Number of samples with data
- SB strand bias at this position
- SOMATIC indicates that the record is a somatic mutation, for cancer genomics
- VALIDATED validated by follow-up experiment
etc. The exact format of each INFO sub-field should be specified in the meta-information (as described above).
Example for an INFO field: DP=154;MQ=52;H2. Keys without corresponding values are allowed in order to indicate group membership (e.g. H2 indicates the SNP is found in HapMap 2). It is not necessary to list all the properties that a site does NOT have, by e.g. H2=0.
Vcf File Viewer Download
Genotype fields
If genotype information is present, then the same types of data must be present for all samples. First a FORMAT field is given specifying the data types and order. This is followed by one field per sample, with the colon-separated data in this field corresponding to the types specified in the format. The first sub-field must always be the genotype (GT).
As with the INFO field, there are several common, reserved keywords that are standards across the community:
- GT genotype, encoded as alleles values separated by either of ”/” or “|”, e.g. The allele values are 0 for the reference allele (what is in the reference sequence), 1 for the first allele listed in ALT, 2 for the second allele list in ALT and so on. For diploid calls examples could be 0/1 or 1|0 etc. For haploid calls, e.g. on Y, male X, mitochondrion, only one allele value should be given. All samples must have GT call information; if a call cannot be made for a sample at a given locus, ”.” must be specified for each missing allele in the GT field (for example ./. for a diploid). The meanings of the separators are:
- / : genotype unphased
- | : genotype phased
- DP read depth at this position for this sample (Integer)
- FT sample genotype filter indicating if this genotype was “called” (similar in concept to the FILTER field). Again, use PASS to indicate that all filters have been passed, a semi-colon separated list of codes for filters that fail, or ”.” to indicate that filters have not been applied. These values should be described in the meta-information in the same way as FILTERs (Alphanumeric String)
- GL : three floating point log10-scaled likelihoods for AA,AB,BB genotypes where A=ref and B=alt; not applicable if site is not biallelic. For example: GT:GL 0/1:-323.03,-99.29,-802.53 (Numeric)
- GQ genotype quality, encoded as a phred quality -10log_10p(genotype call is wrong) (Numeric)
- HQ haplotype qualities, two phred qualities comma separated (Numeric)
If any of the fields is missing, it is replaced with the missing value. For example if the format is GT:GQ:DP:HQ then A|A:.:23:23,34 indicates that GQ is missing. Trailing fields can be dropped (with the exception of the GT field, which should always be present).
Additional Genotype fields can be defined in the meta-information. However, software support for such fields is not guaranteed.
4. Understanding the VCF format and the haplotype representation
VCF records use a single general system for representing genetic variation data composed of:
- Allele: representing single genetic haplotypes (A, T, ATC).
- Genotype: an assignment of alleles for each chromosome of a single named sample at a particular locus.
- VCF record: a record holding all segregating alleles at a locus (as well as genotypes, if appropriate, for multiple individuals containing alleles at that locus).
VCF records use a simple haplotype representation for REF and ALT alleles to describe variant haplotypes at a locus. ALT haplotypes are constructed from the REF haplotype by taking the REF allele bases at the POS in the reference genotype and replacing them with the ALT bases. In essence, the VCF record specifies a-REF-t and the alternative haplotypes are a-ALT-t for each alternative allele.
How do I represent example variation in VCF records?
For example, suppose we are looking at a locus in the genome:
In the above cases, what are the alleles and how would they be represented as a VCF record?
* First is a SNP polymorphism of C/G → { C , G } → C is the reference allele
* Second, 1 base deletion of C → { tC , t } → tC is the reference allele
* Third, 1 base insertion of A → { tC ; tCA } → tC is the reference allele
Suppose I see a the following in a population of individuals and want to represent these three segregating alleles:
How do I represent this? There are three segregating alleles: { tC , tG , t } with a corresponding VCF record:
Now suppose I have this more complex example:
There are actually four segregating alleles: { tCg , tg, t, and tCAg } over bases 2-4. This complex set of allele is represented in VCF as:
Note that in VCF records, the molecular equivalence explicitly listed above in the per-base alignment is discarded, so the actual placement of equivalent g isn’t retained.
For completeness, VCF records are dynamically typed, so whether a VCF record is a SNP, Indel, Mixed, or Reference site depends on the properties of the alleles in the record.
What do example VCF records indicate as variation from the reference?
SNP VCF record
Suppose I receive the following VCF record:
This is a SNP since its only single base substitution and there are only two alleles so I have the two following segregating haplotypes:
Insertion VCF record
Suppose I receive the following VCF record:
This is a insertion since the reference base C is being replaced by C [the reference base] plus three insertion bases TAG. Again there are only two alleles so I have the two following segregating haplotypes:
Deletion VCF record
Suppose I receive the following VCF record:
This is a deletion of two reference bases since the reference allele TCG is being replaced by just the T [the reference base]. Again there are only two alleles so I have the two following segregating haplotypes:
Mixed VCF record for a microsatellite
Suppose I receive the following VCF record:
Vcf File Viewer Android
This is a mixed type record containing a 2 base insertion and a 2 base deletion. There are are three segregating alleles so I have the three following haplotypes:
Vcf File Viewer Iphone
Note that in all of these examples dashes have been added to make the haplotypes clearer but of course the equivalence among bases isn’t provided by the VCF. Technically the following is an equivalent alignment: